
• Research Highlight
X fragile syndrome It is a genetic disorder caused by the gene. FMR1. It is the most common form of inherited intellectual disability and often coexists with other conditions such as autism and epilepsy.
New research has revealed important insights into the genetic mechanisms underlying FXS and related disorders. The study found that the disorders involve extensive silencing of many genes that play key roles in building tissue and properly functioning the brain.
The research was funded by the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke, the Eunice Kennedy Shriver National Institute of Child Health and Human Development and NIH Common Fund 4D Nucleome Program.
How are changes made to the FMR1 Does the gene lead to FXS?
The prevailing theory is that FXS involves changes in FMR1, a gene located on the X chromosome (one of the two sex chromosomes in humans). The first part of FMR1 The gene is made up of repeats of a specific DNA sequence called CGG. The CGG sequence can expand uncontrollably, causing an excessive number of repetitions. a normal length FMR1 The sequence has less than 40 CGG repeats. In contrast, a mutation length FMR1 The sequence has more than 200 CGG repeats.
When the FMR1 CGG reaches this mutation length, causing physical changes in the gene that silence its expression. As a result, FMR1 produces only a small amount or none of its protein, which is necessary for healthy brain function. This is when FXS comes into being. People with FXS who lack the FMR1 protein may experience significant impacts on their development , including intellectual and learning disabilities; speech and language difficulties; and social and behavioral problems, such as hyperactivity and anxiety.
However, the changes in FMR1 alone does not explain the full range of FXS symptoms, as demonstrated by mouse models in which the fmr1 The gene is “deleted” or genetically deactivated. Previous research has also implicated a broader set of genetic mechanisms and gene locations in FMR1 mute.
What did the researchers do in the current study?
Jennifer Phillips-Cremins, Ph.D. , and the study’s first authors, Thomas Malachowski, M.S., Keerthivasan Chandradoss, Ph.D., Ravi Boya, Ph.D., and Linda Zhou, M.D., Ph.D., of the University of Pennsylvania, led the researchers to look beyond the standard FXS model. They examined whether human FXS tissues show changes in the genome (the complete set of genetic material [DNA]) or the epigenome (chemical modifications that influence gene expression without altering the DNA sequence) and whether those changes are specific to FMR1 or also affect other genes.
The researchers combined multiple high-tech analytical methods, including molecular mapping, DNA imaging, epigenetic sequencing, and genetic engineering. Additional computational approaches allowed them to integrate and find patterns in the large data sets they used.
The researchers examined genetic, epigenetic, and imaging data in multiple human cell lines from people with FXS and people without the disorder. They examined the entire genome for changes that occurred on both the X chromosome and non-sex chromosomes (known as autosomes ). Additionally, they worked with the NIH NeuroBrainBank to collect postmortem tissue from an area of the brain linked to the FXS (the caudate nucleus) from people with and without the disorder.
These advanced methods allowed researchers to observe the shape of DNA throughout the genome and how it folds into complex three-dimensional structures within the cell nucleus (known as the 3D genome). Genome folding in 3D space reflects the packaging and interactions of segments of chromatin (the combination of DNA and protein that makes up the genome). Precise folding patterns in the 3D genome are critical for proper gene regulation and cellular function, and deviations from this structure are associated with genetic disorders such as FXS and many cancers.

Across all cell lines and tissues, the researchers compared the three versions of theFMR1 CGG repeat that may occur:
- Normal length (5 to 45 CGG repeats)
- Premutation length (61-199 CGG repeats)
- Mutation length (>200 CGG repeats; FXS)
By analyzing the 3D genome and chromatin changes determined by CGG repeat length, the researchers were able to find epigenetic changes and link them to silencing of gene expression and genome instability that can lead to genetic disorders.
What did the results show?
The results confirmed, in cells and tissues from people with FXS, previously known changes associated with the expansion of the length of the CGG mutation, including epigenetic changes in the FMR1 gene and silencing FMR1 gene expression. The researchers also unexpectedly discovered more widespread changes in DNA.
They found deposits of large pockets of a silencing form of chromatin, known as heterochromatin, which is overfolded and makes the gene less accessible. Heterochromatin pockets extended far beyond FMR1. On the X chromosome, heterochromatin radiated outward to silence genes upstream of FMR1 which are essential for neuronal cell functions, such as circuit connectivity and synaptic plasticity, which could cause learning difficulties commonly experienced by people with FXS. On autosomal chromosomes, heterochromatin silenced multiple genes related to the integrity of skin, tendons, and ligaments, which are tissues clinically affected in people with FXS.
The researchers also found severe DNA folding errors and possible break sites along the DNA sequence or slight expansions of other repetitive sequences within the heterochromatin. These types of higher-order genome folding patterns within cells are critical for proper gene function, so the identified changes in genome structure are likely relevant to FXS beyond local silencing of FMR1.
The Cremins Laboratory coined the term heterochromatin contact-anchored repeat expansion beacons, or BREACHes, to refer to the network of silenced genetic areas they discovered, marked by severe chromatin misfolding and genome instability. The researchers propose that the biological importance of BREACHes is to silence genes involved in processes essential for brain function, including those that control synaptic plasticity that allows the brain to change in response to learning and other dynamic situations. Silencing of these genes may help explain many of the developmental differences observed in people with FXS.
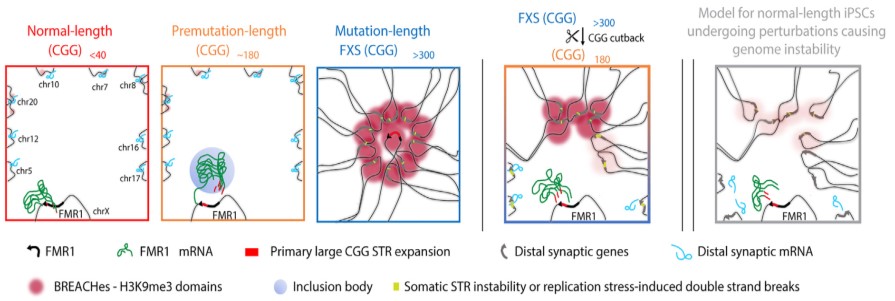
Having established extensive structural and functional changes caused by the length of the mutation FMR1 CGG, the researchers looked at what happened when they reduced the repeat to the length of the premutation. They found that shortening the length of the CGG led to the elimination of heterochromatin in many of the BREACHs. This finding indicates that CGG repeat length is an important contributor to at least some of the severe and widespread 3D genome misfolding and gene silencing seen in FXS and related disorders. It also suggests that reverse engineering the CGG repeat of the mutation length could potentially prevent the emergence of genome-wide defects seen in common genetic disorders in humans.
What do the results mean?
Taken together, the results reveal significant changes in genome structure related to mutation length. FMR1 CGG sequence. These changes, associated with problems in the way DNA folds and instability in the genome, caused genes with important roles in brain function to become silent or inactive, which helped explain many of the symptoms seen in people. with FXS.
The silenced areas encompassed a network of widespread genetic locations that the researchers called BREACHes. BREACHES were observed in multiple cell types and postmortem brain tissue from people with FXS and on both the X chromosome and multiple autosomal chromosomes. BREACHes also included several genes that were not previously linked to FXS, offering new targets for future research.
According to the researchers, the discovery of BREACHes fills an important missing piece in the understanding of FXS. The new finding helps explain the clinical presentation and common symptoms of people with FXS, which previously could not be explained by loss of the FMR1 protein alone. Because genome misfolding occurred not only in FXS cell lines but also in cells with instability in general, BREACHes could be significant for the growing list of disorders marked by unstable repeat expansions.
This study improves understanding of the genetic and epigenetic mechanisms that contribute to FXS disease pathology. The results may, over time, have broad relevance for understanding, diagnosing, and even treating the many disorders characterized by unstable repeat expansions or genome instability, including FXS and cancer.
Reference
Malachowski, T., Chandradoss, K.R., Boya, R., Zhou, L., Cook, AL, Su, C., Pham, K., Haws, S.A., Kim, J.H., Ryu, H.-S., Ge , C., Luppino, JM, Nguyen, SC, Titus, KR, Gong, W., Wallace, O., Joyce, E.F., Wu, H., Rojas, LA and Phillips-Cremins, JE (2023). Spatially coordinated heterochromatinization of long synaptic genes in fragile X syndrome. Cell, 186(26), 5840–5858. https://doi.org/10.1016/j.cell.2023.11.019
Subsidies
MH120269 , MH129957 , DK127405 , DA052715 , HD098015 , NS129317







